**Now Accepting Applications for Pilot Research Project Grants--Read More**
Welcome to our Center!
Our P30 Center at the University of Illinois' Beckman Institute has been funded by the National Institute on Drug Abuse since 2005 and was renewed for another five years until 2024. We provide proteomics, metabolomics, and bioinformatics technologies to biological collaborators at the University of Illinois Urbana-Champaign, and to members of the neuroscience community at other institutions in the United States and throughout the world. Our Center is built around the overarching theme of cell-cell signaling to advance state-of-the-art proteomics / metabolomics technologies focused on the study of addiction mechanisms in the central nervous system.
Why focus on cell-cell signaling? Intercellular signaling plays a crucial role in the organization and coordination of biological systems. A surprisingly large number of physicochemically and structurally distinct molecules are involved in communication among the cells of the brain, with more being discovered each year. These molecules range in size from the small nitric oxide molecule to large >100 kDa heavily post-translationally modified proteins. In addition, these are the endogenous molecules many of the drugs of abuse mimic in terms of receptor binding and other functions. Therefore, these molecules are particularly relevant in drug abuse research and present high-value targets for pharmacological intervention.
Center Structure
Scientific Research Cores
Services are provided to support our collaborators' projects via three synergistic scientific research cores: Sampling & Separation, Molecular Profiling & Characterization, and Bioinformatics, Data Analytics & Predictive Modeling. The services offered by the Neuroproteomics and Neurometabolomics Center on Cell-Cell Signaling are focused on several overarching goals:
Providing metabolomics / peptidomics / proteomics measurement capabilities and bioinformatics services to the Illinois, national and international neuroscience communities working on both fundamental neuroscience research and the study of drug addiction mechanisms.
Discovering the functional roles of metabolites, peptides and proteins in cell-cell signaling, memory, behavior and addiction.
Creating improved MS-based molecular characterization technologies to enable new investigations of cell-cell signaling.
Integrating proteomic and transcriptomic information in support of more accurate molecular identification and molecular network inference using systems biology approaches.
Pilot Research Project Core
In 2019 we added a Pilot Research Project Core to support promising young investigators new to addiction research, and also established researchers interested in exploring new directions in substance abuse research. Are you interested in applying for a pilot research project grant? Read more.
Administrative Core
The Administrative Core offers the support necessary to promote and maintain innovative scientific interactions while facilitating interactions among the three individual research cores and Center collaborators. Are you an established researcher who is interesting in collaborating with us? Read more.
**Center News**
The Analytical Scientist Power List 2023: Jonathan Sweedler, Project Director, voted #3, and Neil Kelleher, Co-Investigator, are both featured in the Category, Leaders and Advocates
NIH Instrument Grant Awarded: Illinois Congresswoman Nikki Budzinski announced on February 14, 2023, that the University of Illinois Urbana-Champaign will receive $1.25 million in federal grant funding from the NIH to purchase a high-end mass spectrometer, which will be used in part to support our NIDA Center's research related to addiction and drug misuse.
The Analytical Scientist Power List 2021:The 100 most influential people in the analytical sciences. Jonathan Sweedler, Project Director, voted #1; Neil Kelleher, Co-Investigator, voted in Top 20
Research News: "Computational Method Provides Faster High-Resolution Mass Spectrometry Imaging"
The Analytical Scientist Power List 2019: The 100 most influential people in the analytical sciences. Jonathan Sweedler, Project Director, voted #1; Neil Kelleher, Co-Investigator, voted #12
Beckman Institute News: "$6M Grant Renews Center That Seeks to Understand the Science of Drug Abuse"
Featured Collaborator
Jim Zaijie Wang is a Distinguished Professor of Pharmacology and Pharmaceutics at the University of Illinois Chicago. His research is focused on the neurobiological and molecular mechanisms underlying chronic pain and drug addiction, and the development of new pharmacological treatments for these conditions. More specifically, his objective is to understand the mechanisms leading to chronic pain, opioid tolerance and addiction, and opioid-induced hyperalgesia.
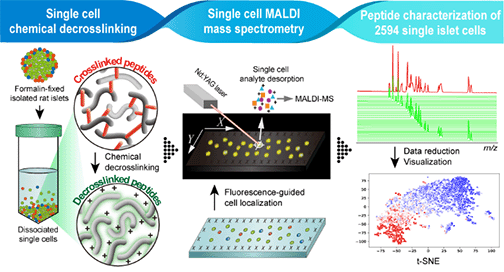
Our initial Pilot Research Project with Wang was centered around hemorphins, an unusual set of opioid peptides formed from the ‘degradation’ of hemoglobin. Interestingly, these are potent antipain peptides. To date there has been no study relating hemorphins (both their specific peptide forms and their levels) to sickle cell disease (SCD), a disease related to changes in the sequence of hemoglobin that can be accompanied by an unusual and debilitating pain phenotype. Our Center performed measurements in plasma and brain tissue to determine whether we could characterize plasma hemorphins from a mouse model of SCD. We implemented a quantitative mass spectrometric approach with the use of multiple reaction monitoring to determine whether hemorphin peptide levels in plasma, specifically LVVH7 and VVH7, are differentially regulated in their mouse model of SCD. In addition to measuring the changes based on sickle cell condition, these two peptides were correlated with each other.
Based on the success of the Pilot project, our effort with Wang is now a full study within our Sampling and Separation and Molecular Characterization Cores. We are quantifying hemorphins in microliter-volume plasma samples and central nervous system tissue of transgenic mice expressing a severe SCD phenotype and correlating this with its associated pain (nociceptive, inflammatory, and neuropathic). Our results indicate that LVVH7 and VVH7 hemorphin levels are reduced in the amygdala but increased in the plasma of our SCD mouse model, suggesting that hemorphin peptides might be involved in mediating pain in SCD.Featured Articles
Multiscale Biochemical Mapping of the Brain through Deep-Learning-Enhanced High-Throughput Mass Spectrometry, Y.R. Xie, D.C. Castro, S.S. Rubakhin, T.J. Trinklein, J.V. Sweedler, F. Lam, Nat. Methods 21, 2024, 521–530. This collaborative project, jointly funded by the National Institute on Drug Abuse, the National Institute on Aging, and the National Institute of General Medical Sciences, has been featured in numerous online articles, including:
- Beckman Institute: Painting a molecular portrait of the brain with mass spectrometry and deep learning
- News-Medical.Net: Deep-learning-enhanced mass spectrometry for biochemical mapping of the brain
- Neuroscience News: 3D Maps Reveal Molecular Complexities of the Brain
- Medical Xpress: Painting a molecular portrait of the brain with mass spectrometry and deep learning
- AZO Life Sciences: The Incredible Impact of Mass Spectrometry on Brain Imaging
- EurekAlert!: Painting a molecular portrait of the brain with mass spectrometry and deep learning
- Newswise.com: Painting a molecular portrait of the brain with mass spectrometry and deep learning
Advances in Multimodal Mass Spectrometry for Single-Cell Analysis and Imaging Enhancement, S.W. Croslow, T.J. Trinklein, J.V. Sweedler, FEBS Lett. 2024, online early view.
Featured Reviews
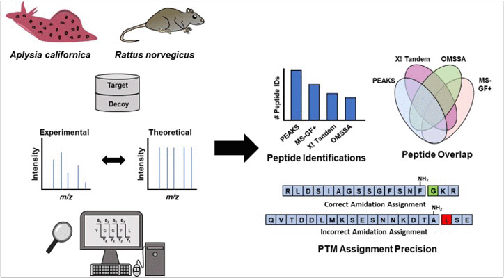
Peptidomics, R. Hellinger, A. Sigurdsson, W. Wu, E.V. Romanova, L. Li, J.V. Sweedler, R.D. Süssmuth, C.W. Gruber, Nat. Rev. Methods Primers 3, 2023, 25.
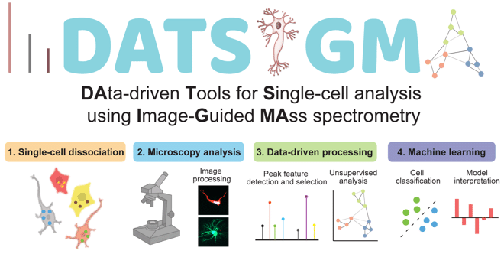
Probe-Based Mass Spectrometry Approaches for Single-Cell and Single-Organelle Measurements, D.C. Castro, P. Chan-Andersen, E.V. Romanova, J.V. Sweedler, Mass Spectrom. Rev., 2023, 1-25 (online early view).
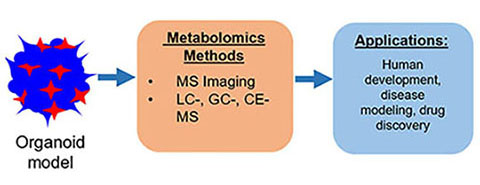
Single Cell Metabolism: Current and Future Trends, A. Ali, S. Davidson, E. Fraenkel, I. Gilmore, T. Hankemeier, J.A. Kirwan, A.N. Lane, I. Lanekoff, M. Larion, L.I. McCall, M. Murphy, J.V. Sweedler, C. Zhu, Metabolomics 18, 2022, 77.