 What do we do?
What do we do?
The Molecular Profiling & Characterization Core characterizes the samples and is at the heart of our Center. Many of the measurements performed by the Core involve specialized mass spectrometers, and we have access to a wide range of instruments. Some of the instruments are located in the laboratories of Neil Kelleher at Northwestern University, while other measurement platforms are on the University of Illinois Urbana-Champaign campus, including an 11 T Fourier transform mass spectrometer, and an ion trap and several time-of-flight instruments, and two mass spectrometry imaging platforms. At Northwestern, Kelleher runs the Proteomics Center of Excellence with a range of high-end instruments available to the Center. These instruments and associated software enable high-performance protein identification and provide unmatched abilities to characterize post-translational modifications.
Due to the importance of determining signal molecule distribution in the brain, mass spectrometric imaging is also being developed using MALDI TOF/TOF technology. This analytical approach provides important clues about molecular function in biological systems, and dramatically eases analysis by uncovering specific brain regions that contain the highest levels of a protein, without requiring microdissection and isolation.
The Molecular Profiling & Characterization Core works closely with the Bioinformatics, Data Analytics and Predictive Modeling Core to increase the throughput and database search specificity of the raw data generated from diverse samples using the various mass spectrometers
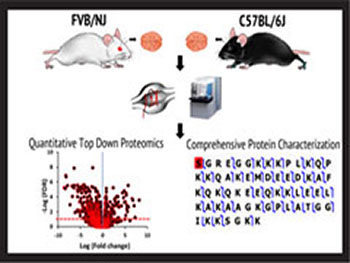
Representative publications
Top-Down Proteomics Enables Comparative Analysis of Brain Proteoforms between Mouse Strains, R.G. Davis, H.-M. Park, K. Kim, J.B. Greer, R.T. Fellers, R.D. LeDuc, E.V. Romanova, S.S. Rubakhin, J.A. Zombeck, C. Wu, P.M. Yau, P. Gao, A.J. van Nispen, S.M. Patrie, P.M. Thomas, J.V. Sweedler, J.S. Rhodes, N.L. Kelleher, Anal. Chem. 90, 2018, 3802–3810. DOI:10.1021/acs.analchem.7b04108
b-Glucocerebrosidase Modulators Promote Dimerization of b-Glucocerebrosidase and Reveal an Allosteric Binding Site, J. Zheng, L. Chen, O.S. Skinner, D. Ysselstein, J. Remis, P. Lansbury, R. Skerlj, M. Mrosek, U. Heunisch, S. Krapp, J. Charrow, M. Schwake, N.L. Kelleher, R.B. Silverman, D. Krainc, J. Am. Chem. Soc. 140, 2018, 5914–5924. DOI:10.1021/jacs.7b13003
Quantitation and Identification of Thousands of Human Proteoforms Below 30 Kda, K.R. Durbin, L. Fornelli, R.T. Fellers, P.F. Doubleday, M. Narita, N.L. Kelleher, J. Proteome Res. 4, 2016, 976–982. DOI:10.1021/acs.jproteome.5b00997
Applying Label-Free Quantitation to Top Down Proteomics, I. Ntai, K. Kim, R.T. Fellers, O.S. Skinner, A.D.t. Smith, B.P. Early, J.P. Savaryn, R.D. LeDuc, P.M. Thomas, N.L. Kelleher, Anal. Chem. 86, 2014, 4961-4968. DOI:10.1021/ac500395k
The C-Score: A Bayesian Framework to Sharply Improve Proteoform Scoring in High-Throughput Top Down Proteomics, R.D. LeDuc, R.T. Fellers, B.P. Early, J.B. Greer, P.M. Thomas, N.L. Kelleher, J. Proteome Res. 13, 2014, 3231-3240. DOI: 10.1021/pr401277r
Comparing Label-free Quantitative Peptidomics Approaches to Characterize Diurnal Variation of Peptides in the Rat Suprachiasmatic Nucleus, B.R. Southey, J.E. Lee, L. Zamdborg, N. Atkins, Jr., J.W. Mitchell, M. Li, M.U. Gillette, N.L. Kelleher, J.V. Sweedler, Anal. Chem., 86, 2014, 443–452. DOI:10.1021/ac4023378
